
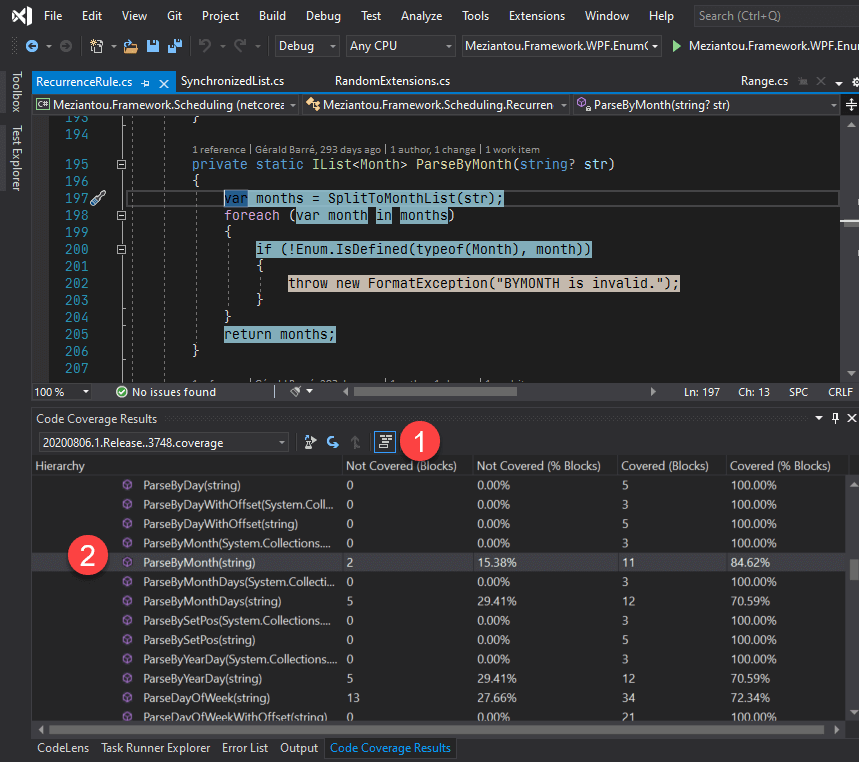
Pairwise alignments of BLAST query and hit sequences showing each HSPįor each hit sequence (horizontal at the bottom), we show how each aligning segment (HSP or High Scoring Pair) aligns to the query sequence (horizontal at the top). This can be more intuitive to understand thatn the somewhat psychedelic colors that NCBI uses by default. A key difference is that in our visualization, darker segments means stronger e-value.
SequenceServer provides a similar overview. SequenceServer’s overview of how hit sequences align to the BLAST search queryĮveryone who has used NCBI’s BLAST will have seen the overview of how hits align to the query sequence.
VISUALIZING CONTACT RESULTS POSTVIEW SOFTWARE
No need to export BLAST results into a different software or to sketch by hand.
VISUALIZING CONTACT RESULTS POSTVIEW DOWNLOAD
Furthermore you can download them in vectorial (SVG) or PNG format, and use them directly for publications. SequenceServer 2.0 provides all of the visualizations below are in every BLAST result. We took inspiration from those efforts so that SequenceServer could better help biologists interpret BLAST results. Visualizations to make BLAST result interpretation easier exist thanks to many efforts, including those at NCBI, those we made for GeneValidator, the work of Nikos Darzentas with Circoletto and the work of Jeff Wintersinger & James Wasmuth with Kablammo. But it’s 2021 - computers should be able to automate that! A picture is worth 1000 words
Speaking from experience, the easiest can be to draw a summary picture on a piece of paper. It quickly becomes a mess trying to integrate large amounts of text and numbers by keeping track of coordinates on the hit sequences, coordinates on the query sequence, and reading frames and DNA strands… This is great for some contexts, but not necessarily, say when you are trying to understand how many times your query gene is present in the target genome. But why should they be? Aligning segments are ordered by biological similarity (e-value). It’s particularly confusing when multiple aligning segments (HSPs) aren’t in chromosomal order. If you have one query with one HSP in a single hit sequence of your BLAST database, interpretation is easy. When running BLAST in the command-line, you end up with a large amount of text. Empowering biologists since 2011 Visualizing and interpreting BLAST alignments


 0 kommentar(er)
0 kommentar(er)
